The tool allows for the retrieval and analysis of GEO Data Sets and GEO Series. GEO Data Sets are processed collections of array experiment results, they are organized into sets of useful information and start with the prefix GDS. Data sets will contain results from experiments into a particular disease state, or from a particular tissue type.
GEO Series are the experimental results as deposited by the researcher. These are generally unprocessed results, subsets of Series can be selectively loaded into SeqExpress.
Help is available on the following topics:
Further information is available about both GEO and SeqExpress.
Both tab separated files and files in SOFT format can be read into SeqExpress using the import tool. To import data select the File->Import option.
Further information/tutorials are available which discuss this process in detail:
Downloading data remotely (from GEO) : Example of reading in both a SAGE and a time series experiment experiment into SeqExpress database from GEO.
Loading data locally: Example of reading the results of a time series experiment into SeqExpress from a local tab separated file.
Loading Annotations: Example of reading/updating LocusLink and Genbank annotations in SeqExpress.
Customising Annotations: Defining how Genbank identifiers for Human (NlaIII) SAGE tags are established within SeqExpress.
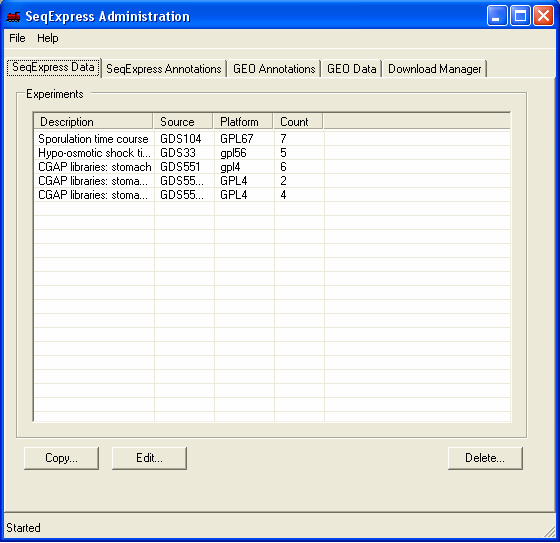
Information about the Experimental results that have been loaded into SeqExpress is available under the SeqExpress Data tab.

Any of the sets of data can be copied, edited (so samples can be added or removed) or deleted.
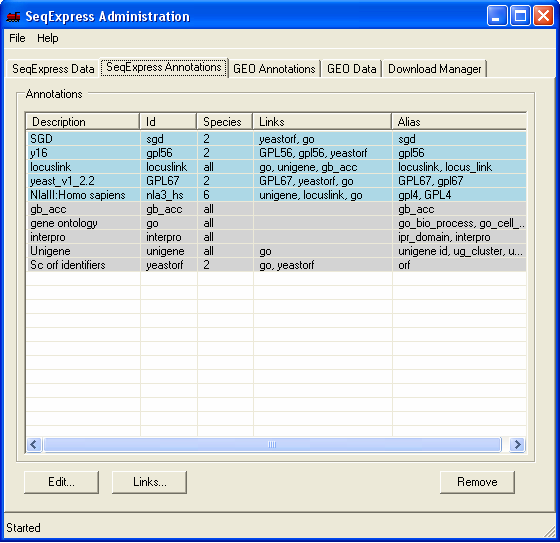
Information about the gene identifiers and any corresponding annotations that have been read into SeqExpress are available under the SeqExpress Annotations tab. Annotation information in SeqExpress consists of:
Additionally information can be provided about:
The Edit button change be used to change the description, the species and add/remove any aliases for the genes or annotations.
The Link button can be used to define any derived annotations and to defined any default annotations.
Annotations and associated information can be removed from SeqExpress using the Remove button.
The Data Sets and Series that are available though GEO are shown under the GEO Data tab.
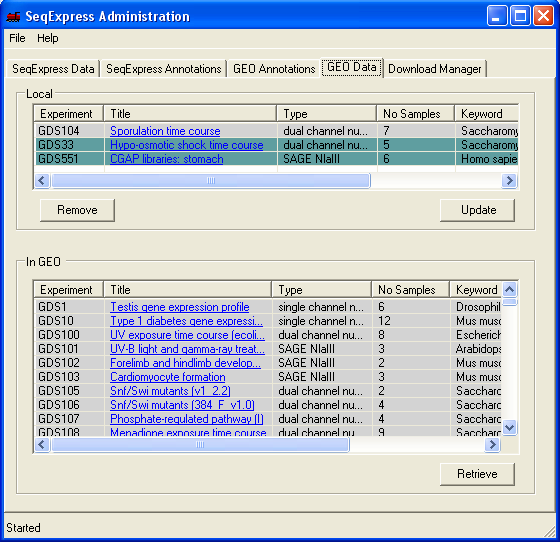
The top table shows the Data Sets or Series that have been loaded into SeqExpress, the bottom table shows the items that are available from GEO. The tables are hyperlinked to the specific GEO information pages. If an item is coloured in gray then it is new, so it has either recently been added to GEO, or has recently been downloaded. An item that is coloured in green is being downloaded.
To retrieve a specific data set from GEO, highlight the item and select Retrieve. The Update button will (re)load a specific data set or series.
Information about the platforms that are available in GEO, and those that have been retrieved are available through the GEO Annotations .tab
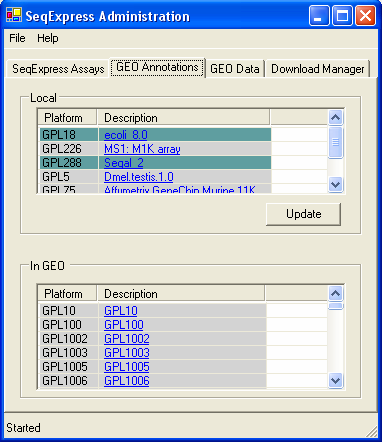
Platforms (and their annotations) that have been downloaded are shown in the top table, the bottom table shows those that are stored in GEO. Items that are gray are new (either recently downloaded, or newly deposited in GEO), those that are in green are currently being added to SeqExpress. Platform and annotation information is automatically downloaded from GEO as it is needed, however annotation information can be reloaded by selecting the Update button (if you wish to change the annotations that have been loaded).
The tables are hyperlinked to the specific GEO Platforms.
The status of the data items that are being retrieved from GEO can be viewed under the Download Manager tab
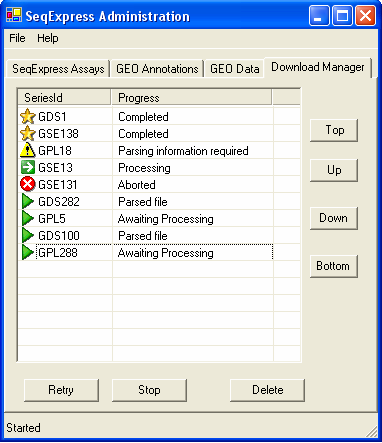
To integrate a data set with SeqExpress the array results must be downloaded, parsed and then loaded into the local SeqExpress database. The download manager provides information about this, the complete process can have any of the following states:
 |
The data has successfully been loaded into SeqExpress, to view the data simply open SeqExpress and select File->Import->"Data set name" | |
 |
Attention is needed, double click on the item and a dialog box will appear. This allows you to choose which items are to be imported. | |
 |
The item is currently being retrieved, parsed or loaded. By default only one item is processed at a time. | |
 |
The item is waiting to be processed. The items are processed in the order they appear in the list, so if an item is moved up the list it will be processed sooner. | |
 |
There has been a problem with processing the item, this is typically due to either a network error or because the Stop button has been pressed. The Retry button will cause the tool to (re) start the whole retrieval, parse and load procedure. |
The delete button will remove the selected item from the list. An item that has been Completed can safely be deleted from the list.
It is possible to customise the behaviour of the tool and to specify if a server proxy should be used. To specify a proxy server select the File->Proxy Settings... option. To enter the details you first need to select the Use Proxy box. Once the details have been enter select OK.
By selecting the File->Properties option two aspects of the GEOTool can be customised:
The tool has the standard advantages and disadvantages of using an underlying DBMS, as compared to an indexed flat file system. The system is transactional and robust, however maintance of indexes, large scale updates/deletes and referential integrity all mean that the system can become unresponsive if it is overloaded with multiple requests. The maintance scripts are run in a nonexclusive manner, so the system will always be available, that is to say tasks such as rebuilding indexes do not cause block/wait on the database. Such non-locking behaviour means that the indexing takes a longer time to perform.Upon starting the tool, an integrity tests are performed, any items that fail the test are marked for deletion.
To ensure a high level of responsiveness bulk loads (bcp) are used. Additionally, cascade deletes are not performed on the database; instead items are marked for deletion, so that they can be deleted using small atomic transactions (so that the transaction log does not become too large to handle on a standard desktop machine).. As the deletion is an ongoing process that runs in the background, it is possible that if experiments are constantly added/deleted then the maintance procedures will not be able to keep pace with the changing database. If the system becomes unresponsive, leave the tool running on an unaltered database until the maintance scripts have finished (the status of the maintance scripts can be viewed using the monitor tool available from the file menu, you should select the database maintance thread to view the current maintance status).