|
Downloading data remotely (from GEO) | ||||||
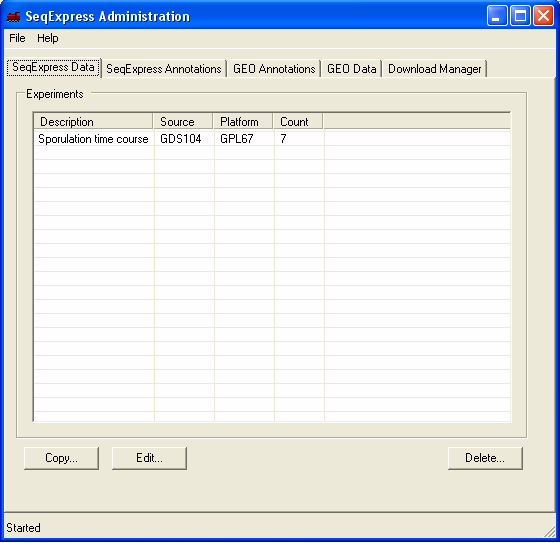
Once SeqExpress has been installed you should start the GEO Tool, from Start->Programs->SeqExpress->GEOTool. Depending on the server version, when the tool is first started a notice may appear informing you that an update is occuring. Additionally once updated a preliminary maintance check is started, it is recommended (but not essential) that you always run the check when you start the tool. When the tool starts you will be shown information about: SeqExpress Data: what experiments are currently loaded into SeqExpress. SeqExpress Annoations: what (gene) annotations are currently loaded into SeqExpress. GEO Annotations: what annotations (for the various array platforms) are available from GEO. GEO Data: what experiments are available from GEO. Download Manager: what files are currently being retrieved, parsed or loaded into SeqExpress By default one set of experimental results is loaded into SeqExpress, as well as annotations from a variety of databases (SGD, LocusLink, Human SAGE tags, GO term etc.)
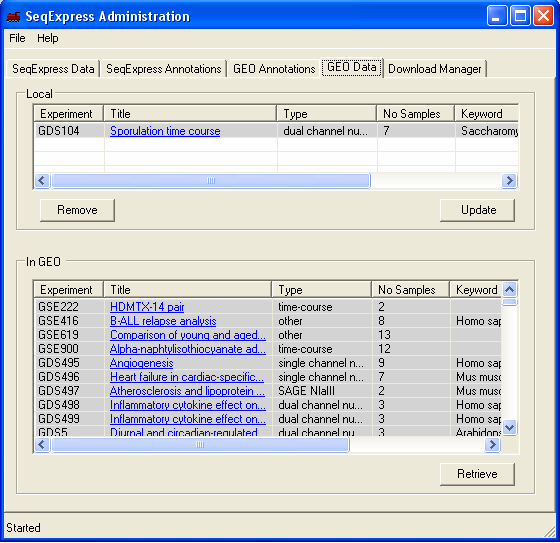
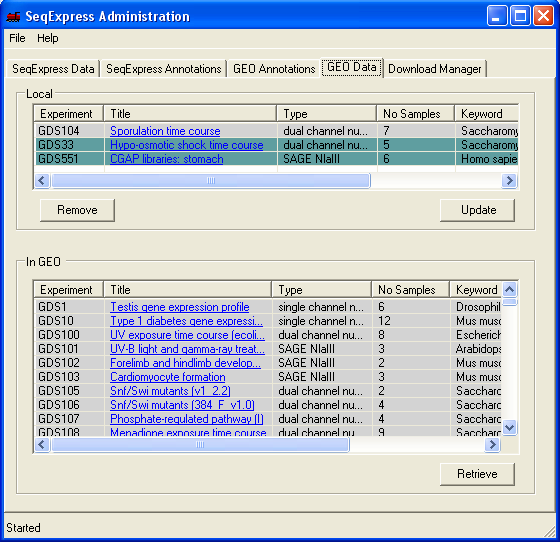
To request for assay information to be loaded into SeqExpress, select the 'GEO Data' tab and choose the required series or data set from the bottom list and press the retrieve button (in this example below GDS551 and GDS33 are retrieved).
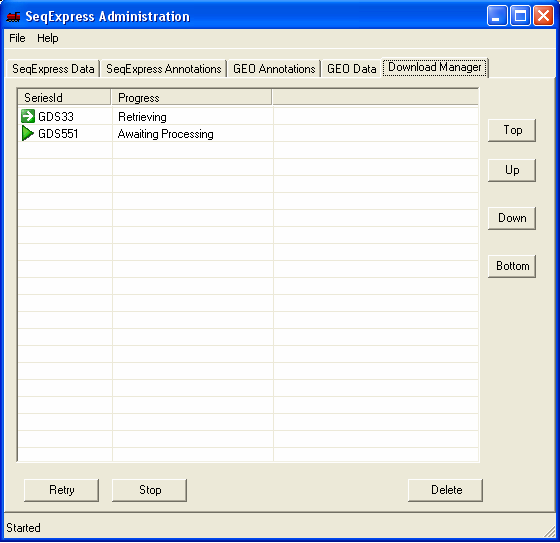
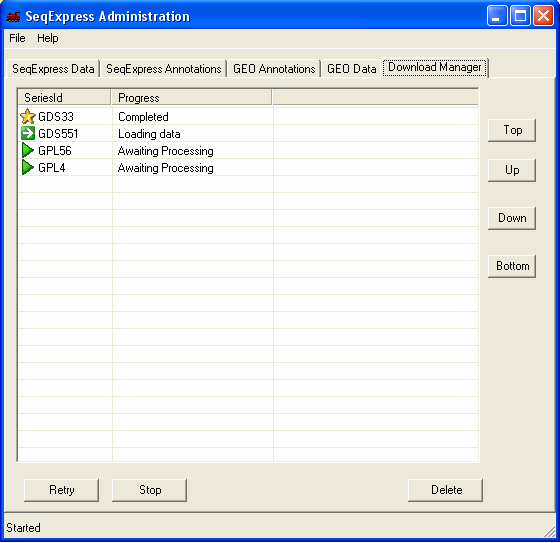
If you select the 'Download Manager' tab you can see the status of the downloads. When downloading data sets from GEO the import is fully automatic, when downloading series information you choose which items of information you wish to import. The two GEO data set assays that are being imported are a SAGE study on stomach tissue (GDS551) and a yeast time series study (GDS33).
The tool will download, parse and then load the information from GEO. If further information is needed (for example an associated annotation/platform file), then this is retrieved automatically.
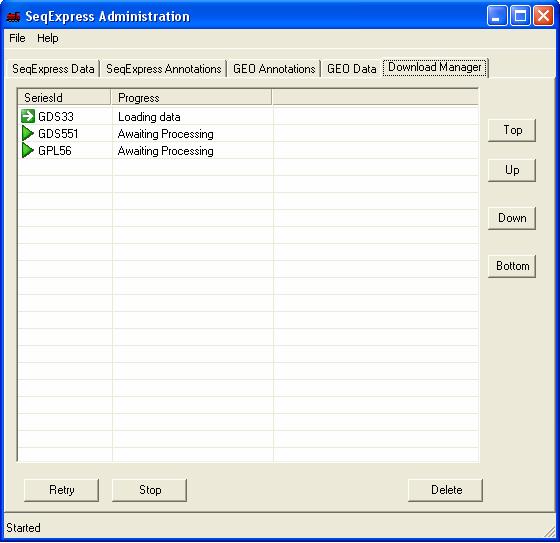
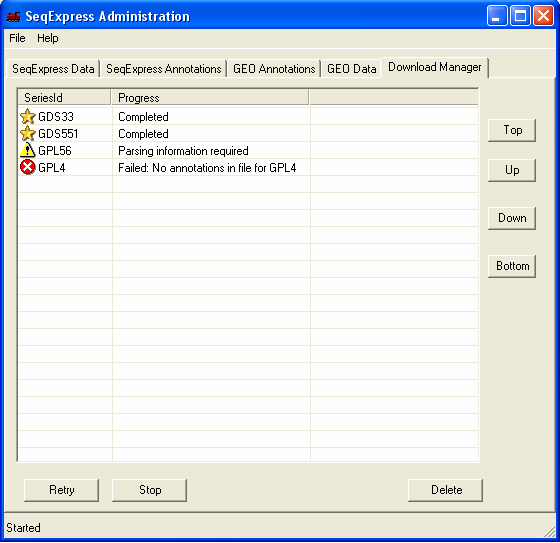
A 'gold star' shows when data has been successfully retrieved from GEO. Once the information has been loaded it can be viewed in SeqExpress by selecting File->Import
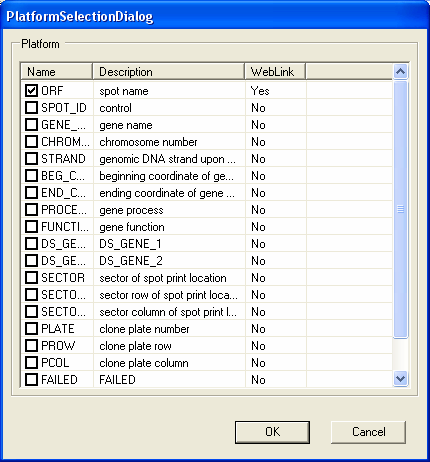
If there is an error in the download this is shown by a 'red cross'. The error that occurs with the GPL4 platform is due to no annotations being available (in this case it doesn't matter as the latest human NauIII SAGE tag unigene mappings have already been loaded into the database). If a choice about what data to import is required this is shown by a 'yellow exclamation mark' - to make a choice simple double click on the item. Most of the choices have default options which can generally just be accepted. In this case you should double click on the GPL56 item to select which annotations should be imported.
To make a choice as to which data to import double click on the item. A dialog will appear allow you to select the data items to be imported (in GEO series this will be the columns of data, and for GEO Platforms this will be the annotations). In this case the default options are fine, so you should select the OK button to continue the import procedure.
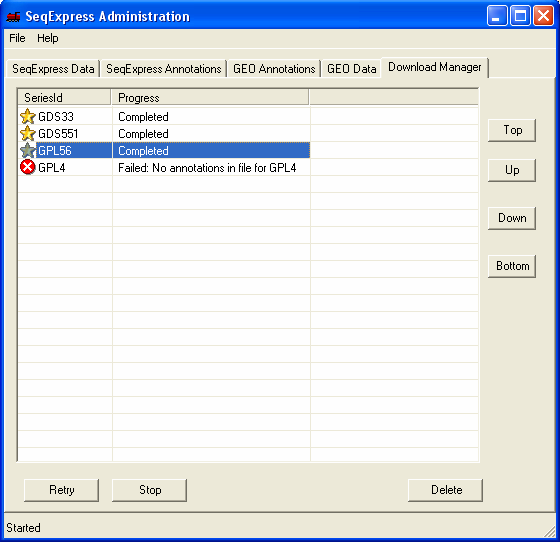
Once you have made your choices, the process will continue.
The currently loaded data items can be manipulated in the other tabs. Colour coding is used to show items that have recently been added to GEO. Once imported that data can be analysed directly from inside SeqExpress.
Any defects found in the tool should be reported by email bugs@seqexpress.com. More help is available.
|