|
Loading data locally | ||||||
To load local proprietary data into SeqExpress you should select File->Import... in the GEOTool and choose the file containing the experiment data. SeqExpress supports the import of either tab separated formatted files of GEO SOFT formatted files. In this example, a file containing tab separated expression results is imported into SeqExpress. Tthe preferred format for importing data is SOFT, and should be used whenever possible (as information about the platform, species etc can be explicitly defined). In this example we are importing information from one of the Spellman cell cycle experiments.
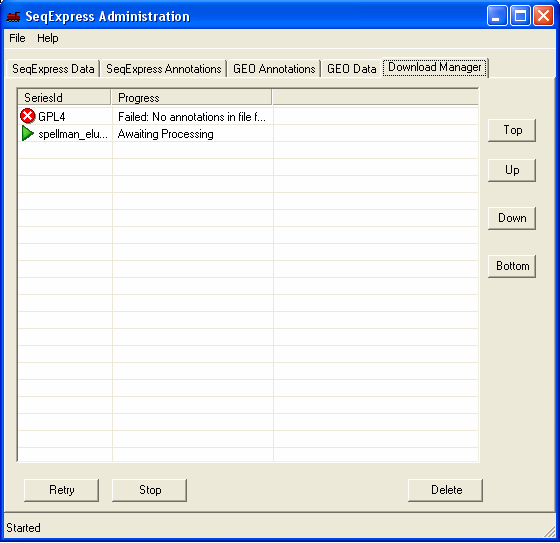
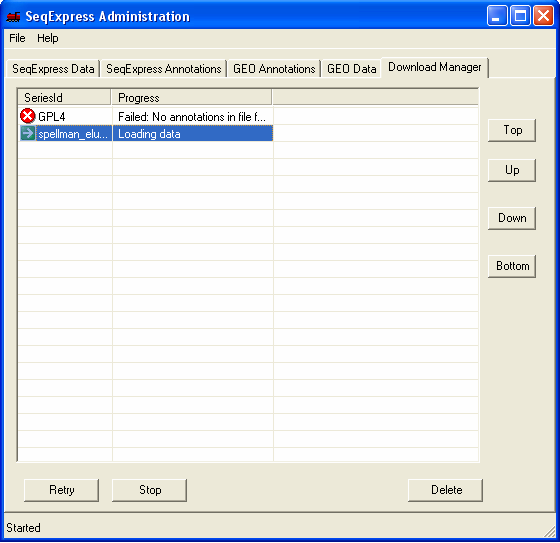
Once the file has been chosen it is queued for importing. You can view the queued items by selecting the Download Manager tab, the higher the item is in the list the higher its priority (so the sooner it will be imported).
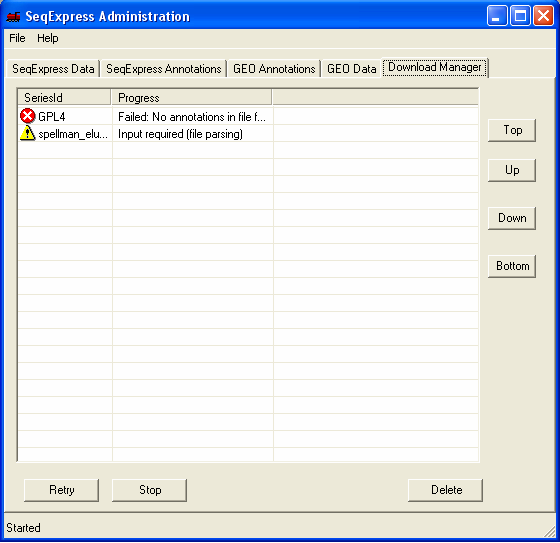
Within a short time the icon will show that the item which is being imported requires futher input. You should double click the item to enter the parsing and loading information.
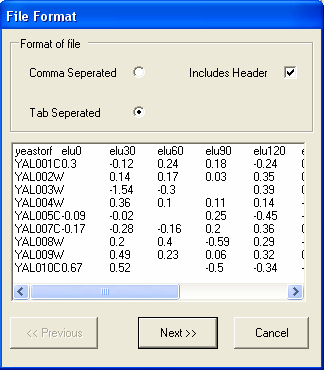
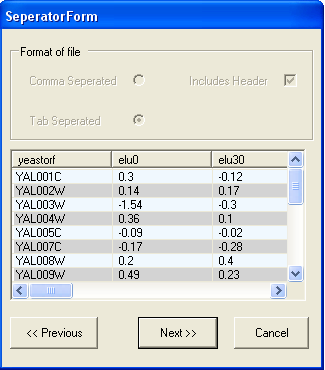
In this example the file is tab separated and has a header line, so the default options are appropriate. You should select the Next button.
The next screen shows that the file is correctly formatted, to continue select the Next button.
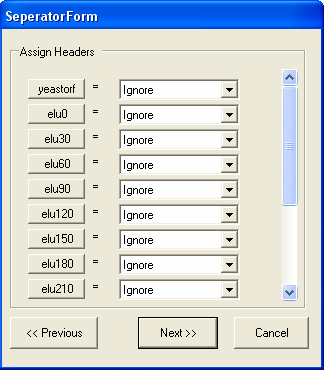
The types of data in each column need to be specified. For importing tab separated files 5 types of column type are supported:
By default all the columns are ignored.
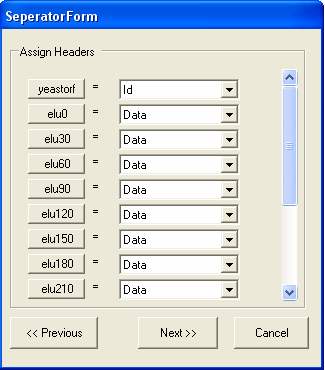
In this case the first column contains the ID for the gene, and all the other columns contain data. It is possible to change the name of the column by clicking the corresponding column button, in this case this is not needed. The name of the column corresponds to the type of ID that is being imported. The image below shows that the ID column contains 'yeastorf' identifiers and is the first column, all the other columns contain experimental data. You should select NEXT when you have filed in the form.
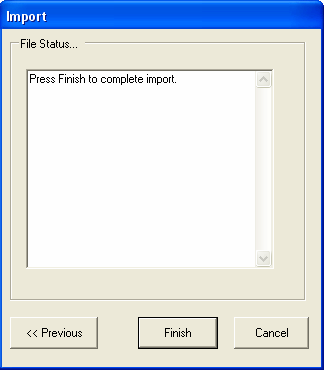
Selecting FINISH will start the import procedure, any errors that occur will be reported in the text box (for example if you are importing experiment data and the Ids are not unique).
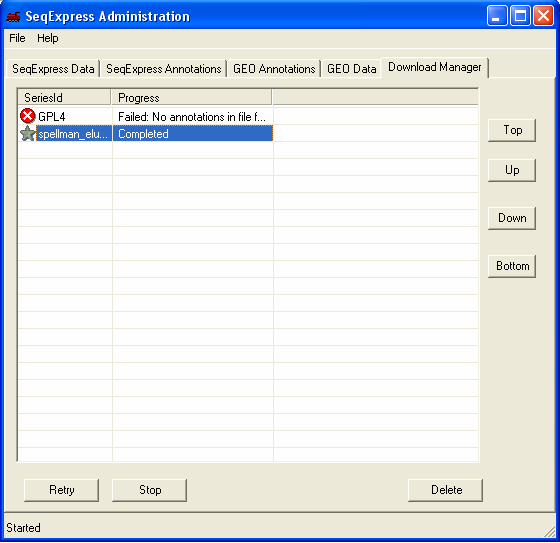
The file will be loaded into SeqExpress, this will take a few minutes.
When the load is complete a 'gold star' icon is shown.
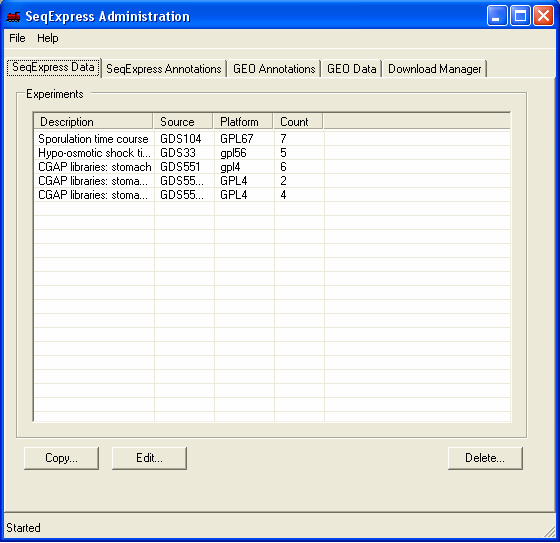
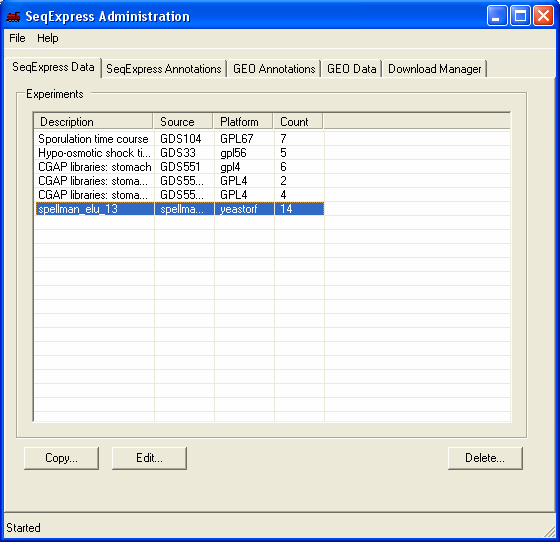
The information in the file is now loaded into SeqExpress, and can be opened using the File->Import menu. The available experiment data can be deleted from the SeqExpress server using the Delete button under the SeqExpress Data tab.
|