|
Loading Annotations | ||||||
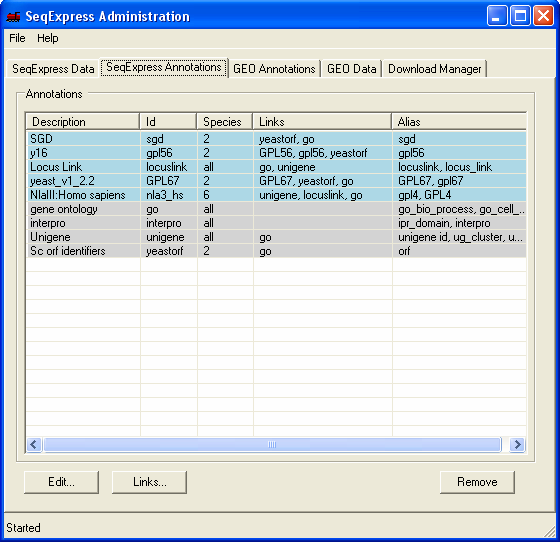
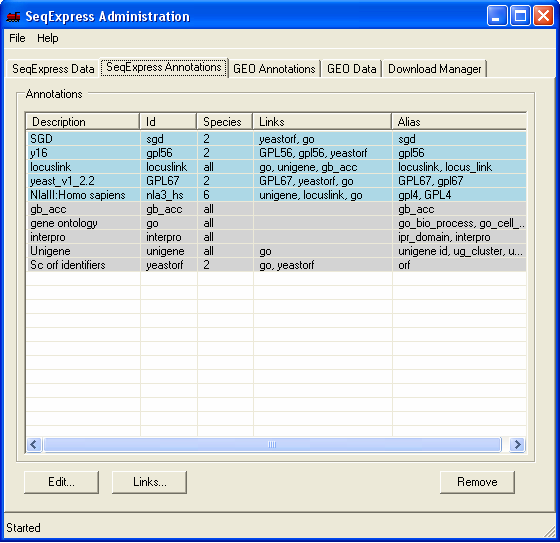
In this example Genbank Identifiers for all locuslink entries will be imported, this is done by importing information directly from the NCBI locuslink to GI mapping file. Once loaded, it will be possible to directly lookup individual genbank entries from a corresponding locuslink entry. The currently loaded annotations can be viewed by selecting the SeqExpress Annotations tab.This shows the:
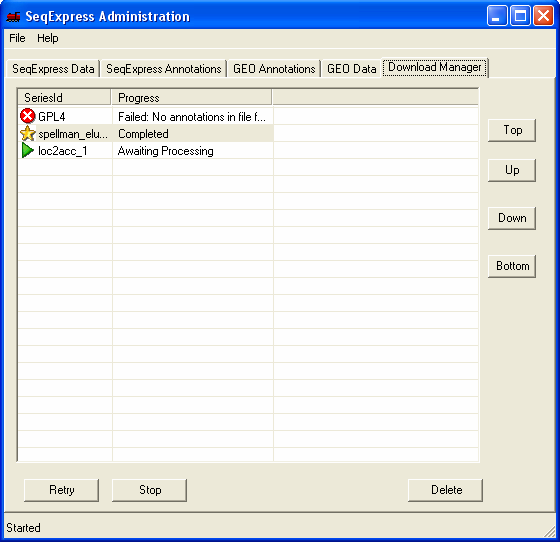
To start the import you should download the mapping file from NCBI , and then select it using the File->Import... menu option. The annotation file will then be queued for import, the current state of the importation can be viewed under the Download Manager tab.
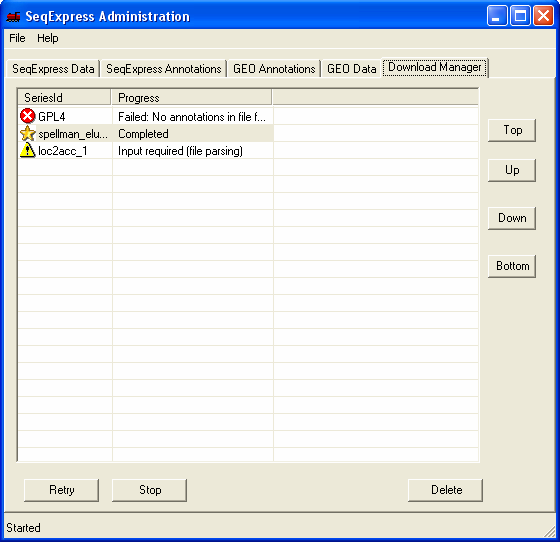
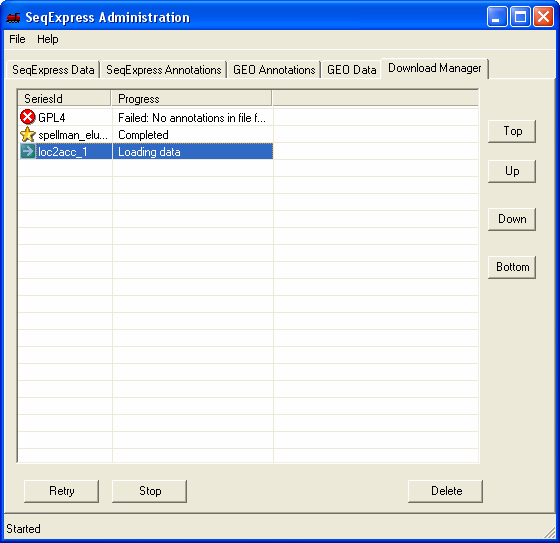
To complete the import og the annotaions further information is required, this is shown by the' yellow exclamation mark' icon. You should double click the item to enter the required information.
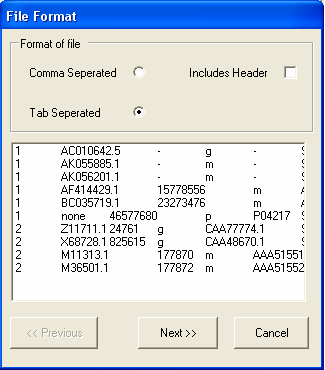
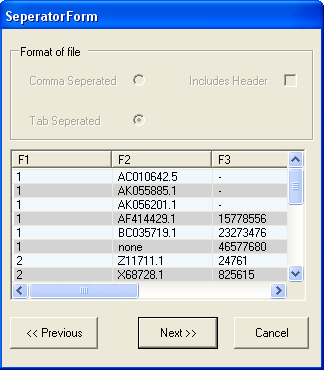
In this example the file is tab separated, but does not contain a header line (so the includes header check box should be unselected). Select Next to continue.
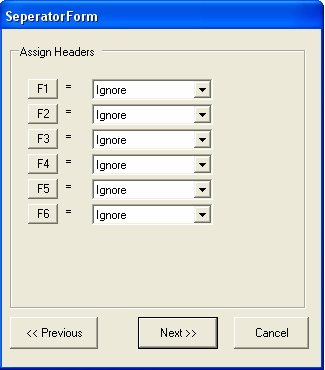
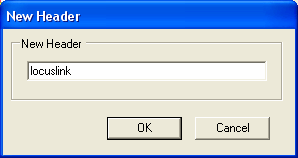
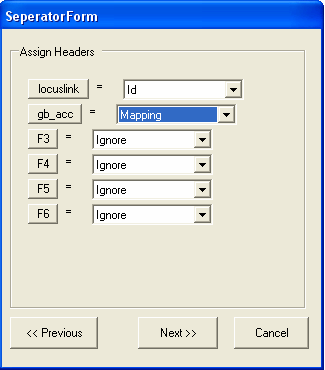
The file is formatted correctly, so you should select the Next button. Information about the type of each column needs to be specified. As only annotation information is being imported, it does not matter if the Id column is unique.The first column contains LocusLink identifiers, so we need to chang the name of the column from F1 to 'locuslink' (we can alternatively use one of the aliases in the SeqExpress Annotations tab that have been defined for locuslink).To change the name of the column select the 'F1' button.
Now change the name to locuslink, and select OK
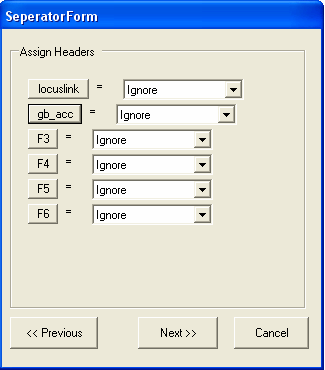
We should also change the name of the second column to gb_acc or similiar. As this is the first set of genbank annotations have been read into SeqExpress, this identifier will be used to refer to this type of annotation from now onwards (alias information can be defined for gb_acc after it has been imported, commonly used aliases include GB, GenBank,GI) To change the name select the F2 button, enter gb_acc and select OK.
Now we specify that the first column contains IDs and the second column contains annotation mappings. Additionally textual descriptions for the IDs (in this case locuslink) could also be imported. Select Next to continue.
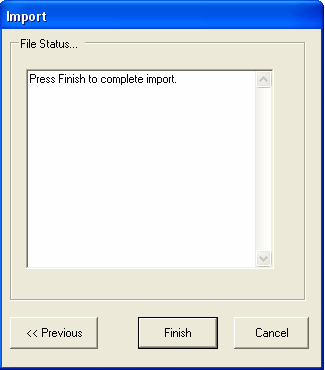
Select Finish to complete the parsing of the file, any errors that occur will be reported in the text box.
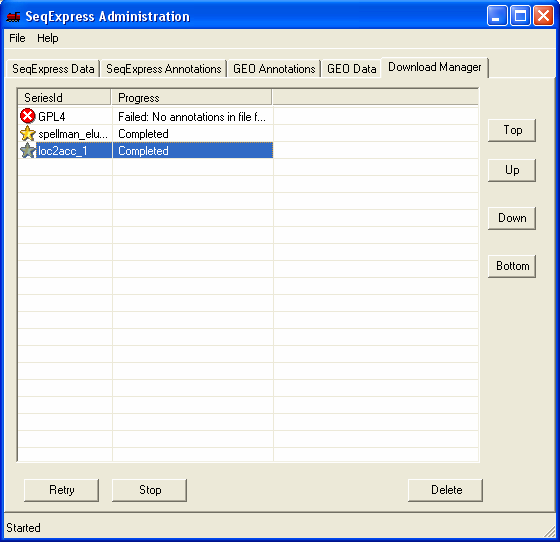
The Download Manager tab shows that the annotation data is being loaded, this will take approximately 5 minutes (depending on machine performance). Approximately 300,000 annotations are being imported.
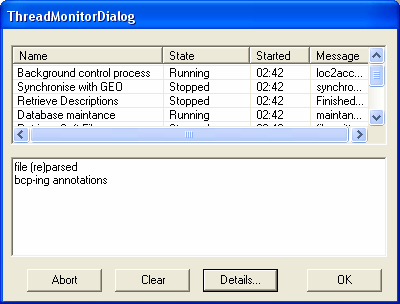
A monitor tool is available which allows you to view the current state of any subprocesses that are running in SeqExpress (also provides any error/status information). This can be accessed from the File->Monitor.. menu. This tool is for general interest, and is not needed to use SeqExpress.
Once completed the 'gold star' icon is shown.
The newly imported locuslink to gb_acc mappings have been entered into SeqExpress (so it is now possible to navigate from locuslink entries to the corresponding genbank entries). This can be seen as the locuslink 'links' column now contains the gb_acc link. The next tutorial shows how mappings between the different identifiers can be customised.
|