A number of different search options are available, these will create a selection of genes and will be added to the Gene tab in the main SeqExpress window.
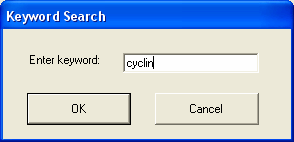
By selecting the keyword option in the edit menu it is possible to find all genes/tags that contain the keyword. This is done by search the textual descriptions of each of the default annotations associated with the tag (so if the mapping is changed then the results will change). The keyword dialog can be used to enter the search term.

The keyword search is for a substring, so in the above query all genes that contain 'cyclin' will be returned.
Values above and below certain ranges within either on or all of the experiments can be selected. The filter can also be used to specify all genes that are above or below one or two standard deviations from the mean.
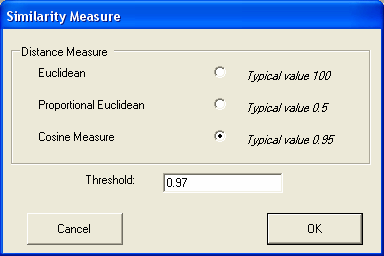
Techniques are provided to measure the similarities between the gene expression profiles in the different experiments Genes can be found with similiar expression profiles (with similiarty based on either cosine or eucledian distance measures).
Using the 'what-if' based tool a profile can be defined using the sliders and then number of items that match that profile are shown in the bottom left hand side of the dialog. The profile is defined as a series of greater or less than values for each experiment, so it is possible to conveniently to find out then number of items (and then select them) that have high expression in a number of experiments, but have low expression in others (e.g. disease/non disease states). The "add selection" button will add the currently selected items as anew selection in the gene tab.
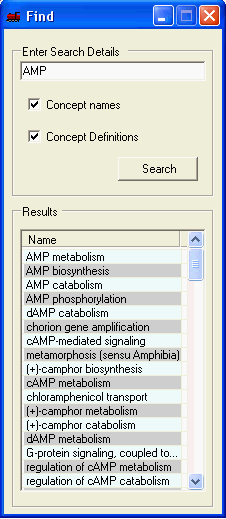
Searching through the currently selected ontology concepts and their definitions is supported. The results are ordered by the 'exactness' of the match. To perform a search simply enter the search term, select whether to search the concept names and/or their definition and select search. Selecting a term from the result list will select all relevant genes within all open visualisations and the corresponding term in the function tab.
Selected Edit->Copy Selected will copy all genes which have been annotated with the specific ontology term to be added as a new selection in the gene tab.