Two tools are provided for viewing and editing sets of genes:
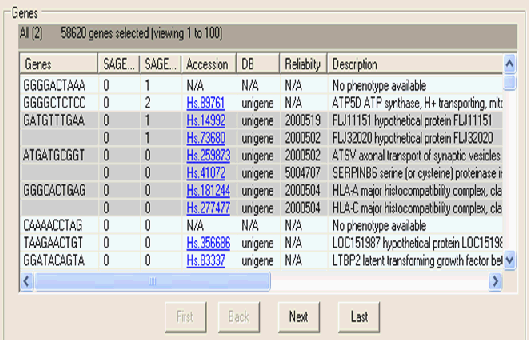
A Gene Table gives information about genes, their individual experiment expression value, associated annotations and any descriptions
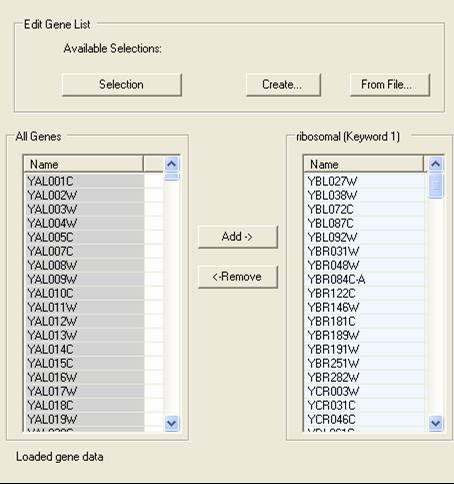
A Gene List Editor allows for the editing of sets of genes.
Detailed information about the genes that were analysed within the experiment are viewed through the Genes table, the genes table shows:
The unique identifier for the gene (this could be the well number, an internal identifier, the sage primer sequence etc…)
The individual experiment details: he results for the gene within each experiment are shown in separate columns. If null values are stored these are defaulted to zero.
Accession id: if there are any annotations available for the gene then this information is shown. If available a hyperlink to an external data source is provided.
DB: the name of the databank which the annotation comes from.
Reliability: if multiple phenotypes are mapped to a single gene, if available a score is given showing the reliability of the suggested phenotype(s)
Description: A text description of the phenotype function is given.
By using the Edit->Annotation option it is possible to select a different default annotation databank. If more than one annotations is mapped to the gene then the list of the annotations are shown with a reliability score (if available). The annotation information is retrieved on the fly from the server, this means that there will be a delay as information is retrieved in chunks.

Navigation buttons at the bottom of the Genes table allow for paging through the subset of genes. Items that are selected within this table can be saved as separate collections, this is done by using the 'copy selected' item in the Edit menu.
It is possible to edit the individual genes in a selection by using the Gene List Editor option. This allows for the direct addition or removal of genes from a specific group. By using the Selection button at the top of the editor it is possible to choose the selection to be edited. The genes that are in the chosen selection are shown on the right. Genes are added or removed from the selections using the controls at the bottom of the dialog. A new selection can be created by select the Create button, and list of gene identifiers in an external file can used to generate a selection by using the From File... button..